Focus
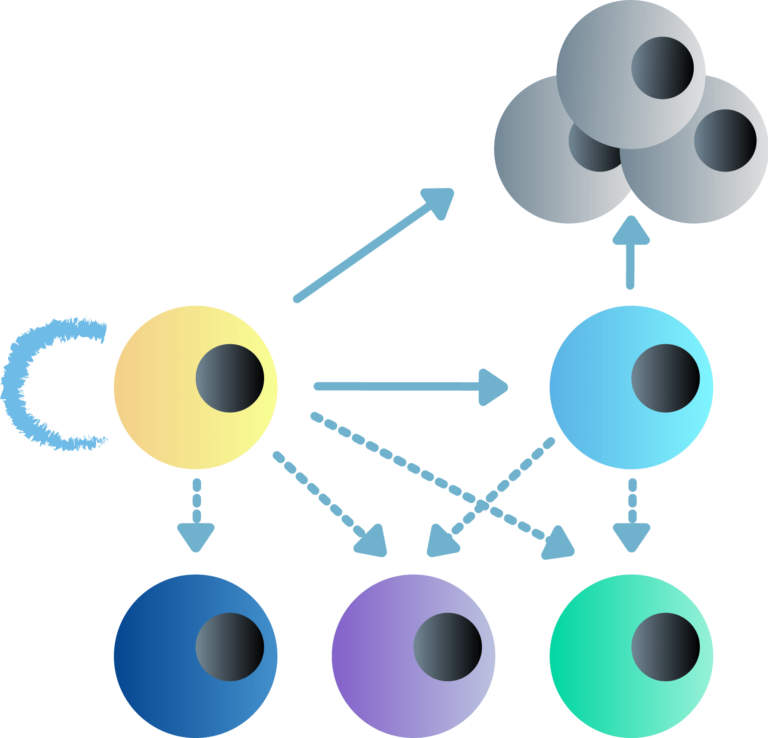
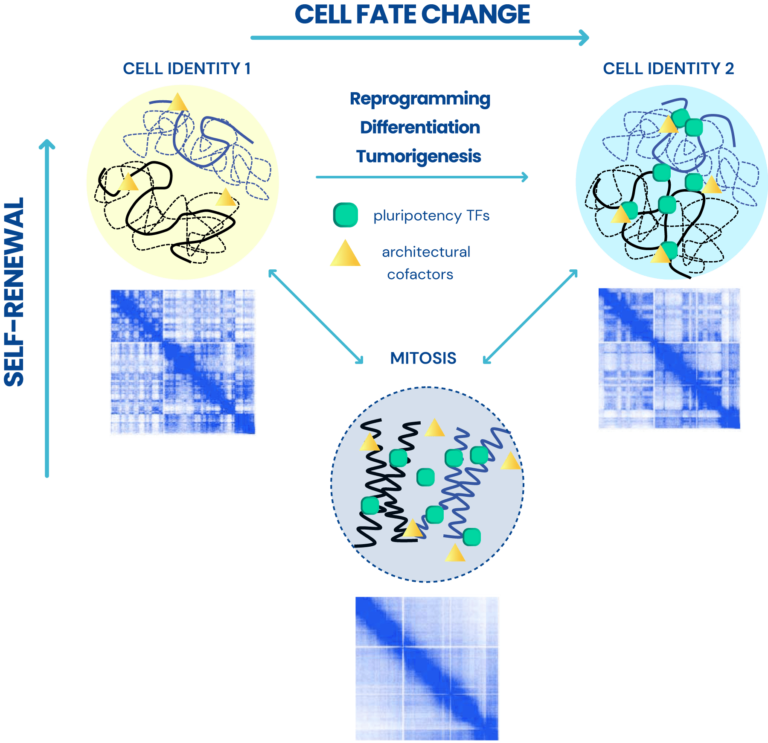
Our main research focus is to understand the complex functional interplay between transcription and 3D chromatin architecture and their impact on cell fate decisions, such as reprogramming, self-renewal or differentiation, and tumorigenesis. Specifically, we have set two key research directions: (A) To establish a computational and experimental platform for the identification and functional interrogation of core regulatory elements and factors that control cell fate decisions in embryonic development and various disease contexts. (B) To determine mechanisms of epigenetic inheritance of cell identity and exploit temporal windows of vulnerability of cell fate.
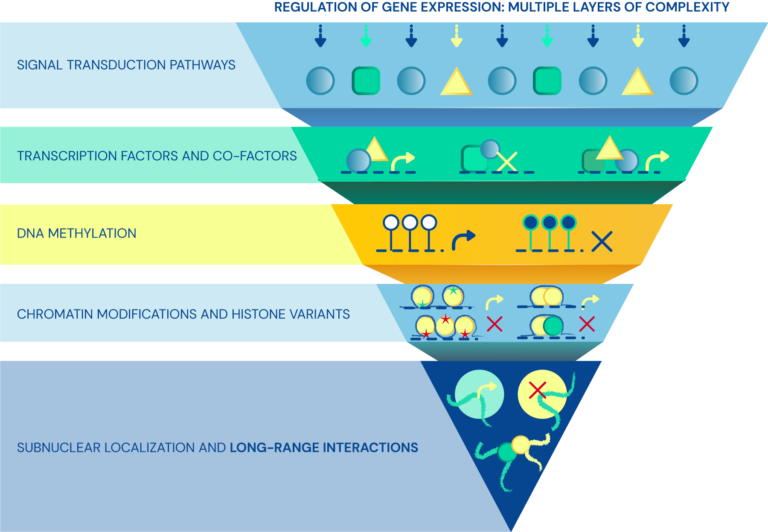
Regulatory layers of cell-type specific programs
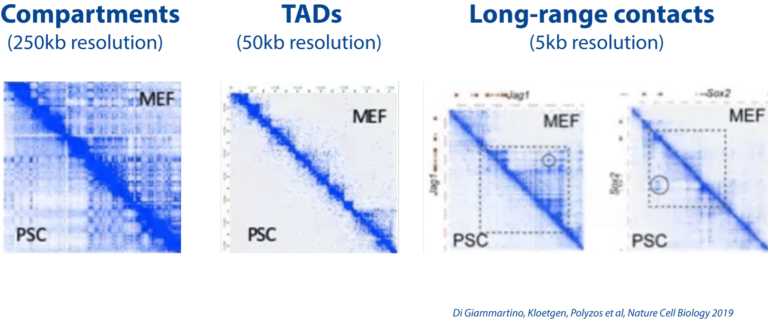
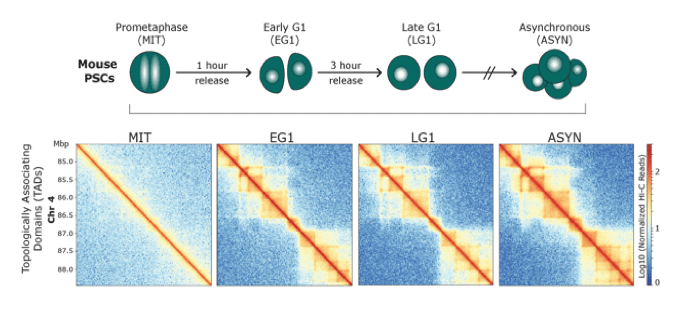
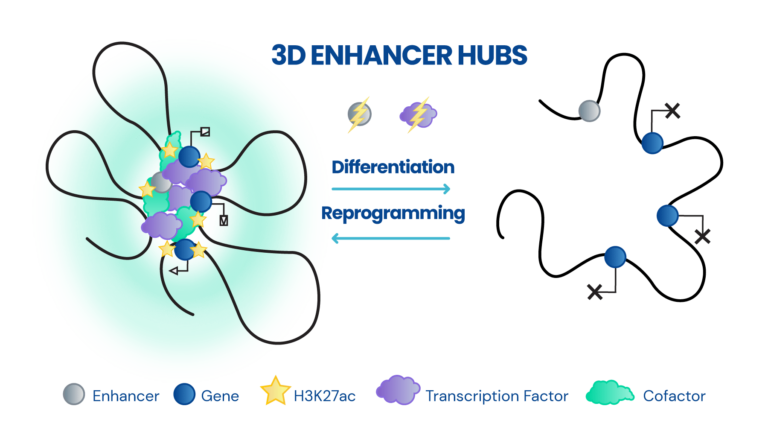
Dissecting the role of 3D chromatin (re)organization in cell fate decisions
Approach
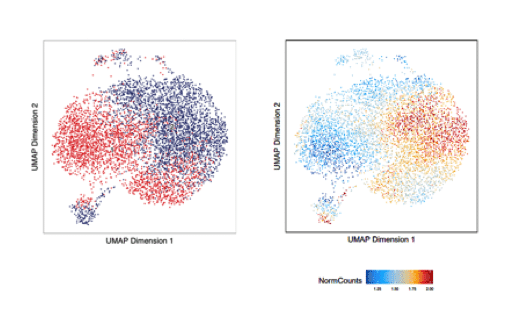
We utilize a variety of dynamic cellular systems, including iPSC reprogramming, differentiation, immune response, tumorigenesis and cell cycle to build 4D molecular roadmaps and address our key questions. We use high-throughput sequencing technologies such as 4C-Seq, HiC, HiChIP, ChIP-seq and PRO-seq and integrative computational analysis to dissect regulatory mechanisms of cell type-specific gene expression programs. We also employ advanced CRISPR/(d)Cas9-based genetic and epigenetic approaches for precise modulation and functional testing of our hypotheses. Finally, we are expanding to single-cell technologies to interrogate functional heterogeneity and rare stem-like populations in vivo and ex vivo.
Projects / Directions
- Constructing 3D enhancer-promoter networks in early development and disease in order to identify and target core regulatory elements
-
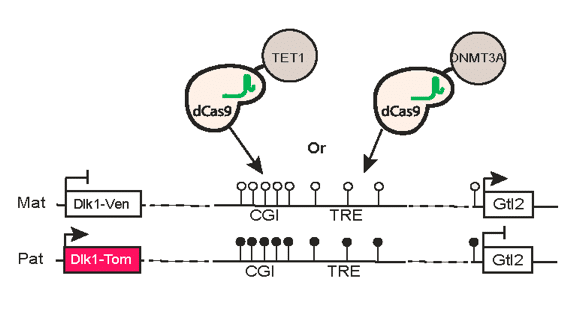
Dissecting epigenetic mechanisms of cellular plasticity using cellular reprogramming.
-
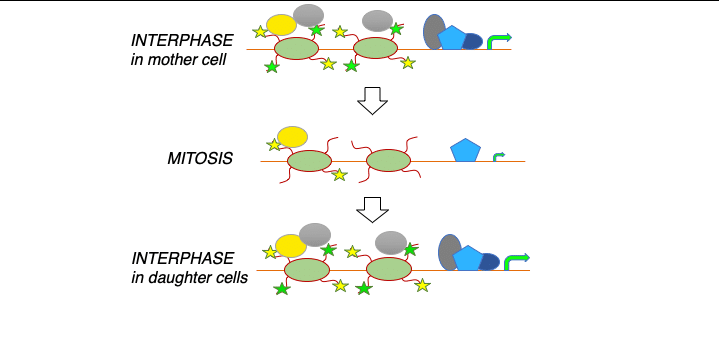
Identifying principles and critical regulators of cell fate inheritance after cell division
- Characterizing stem-like subpopulations and epigenetic features that contribute to lymphomagenesis or lymphoma repopulation