Selected publications
-
T follicular helper cells transiently unlock a plasticity state in germinal centre B cells during the humoral immune response.
Scourzic L*, Izzo F, Teater M, Polyzos AP, Cucereavii L, Chin CR, Papin A, Pinto HB, Mlynarczyk C, Tsialta I, Xia M, Lidoski A, Myers RM, Israel EM, Venturutti L, Mackay SP, Hoehn KB, Skoultchi AI, Béguelin W, Stadtfeld M, Chen Z, Landau DA, Melnick AM* and Apostolou E*. Nature Cell Biology 2026 PMID: 41466145
-
Three-dimensional regulatory hubs support oncogenic programs in glioblastoma
Breves SL, Di Giammartino DC, Nicholson J, Cirigliano S, Mahmood SR, Lee UJ, Martinez-Fundichely A, Jungverdorben J, Singhania R, Rajkumar S, Kirou R, Studer L, Khurana E, Polyzos A, Fine HA and Apostolou E. Molecular Cell 2025 PMID: 40147440
-
Coordinated repression of totipotency-associated gene loci by histone methyltransferase EHMT2 via LINE1 regulatory elements.
Chatterjee K*, Uyehara CM*, Kasliwal K, Madhuranath S, Scourzic L, Polyzos A, Apostolou E* and Stadtfeld M* EMBO Reports 2025 PMID: 41366101
-
MED1 IDR deacetylation controls stress responsive genes through RNA Pol II recruitment.
Lin R, Mo Y, Barrows D, Mei W, Onikubo T, Sun J, Zhang Z, Apostolou E, Tavazoie SF, Roeder RG. Nature Chemical Biology 2025 PMID: 41131199
-
Enhancer remodeling by OTX2 directs specification and patterning of mammalian definitive endoderm
Ee LS, Medina-Cano D, Goetzler E, Uyehara C, Schwarz C, Salataj E, Madhuranath S, Evans T, Hadjantonakis AK, Apostolou E, Polyzos A, Vierbuchen T, Stadtfeld M. Developmental Cell 2025 PMID: 40834858
-
Reprogramming Stars #23: Charting Cell Fate Crossroads from the Interplay Between Epigenetics, Transcription, and 3D Chromatin Architecture—An Interview with Dr. Effie Apostolou
-
3D chromatin hubs as regulatory units of identity and survival in human acute leukemia
Gambi G, Boccalatte F, Rodriguez Hernaez J, Lin Z, Nadorp B, Polyzos A, Tan J, Avrampou K, Inghirami G, Kentsis A, Apostolou E, Aifantis I, Tsirigos A. Molecular Cell. 2025; PMID: 39719705.
-
Single-cell analysis of bidirectional reprogramming between early embryonic states identify mechanisms of differential lineage plasticities in mice
PMID: 39729987
-
DDX18 coordinates nucleolus phase separation and nuclear organization to control the pluripotency of human embryonic stem cells
Shi X, Li Y, Zhou H, Hou X, Yang J, Malik V, Faiola F, Ding J, Bao X, Modic M, Zhang W, Chen L, Mahmood SR, Apostolou E, Yang FC, Xu M, Xie W, Huang X, Chen Y and Wang J. Nature Communications 2024; PMID: 39738032
-
ChromaFold predicts the 3D contact map from single-cell chromatin accessibility
Gao VR, Yang R, Das A, Luo R, Luo H, McNally DR, Karagiannidis I, Rivas MA, Wang ZM, Barisic D, Karbalayghareh A, Wong W, Zhan YA, Chin CR, Noble WS, Bilmes JA, Apostolou E, Kharas MG, Béguelin W, Viny AD, Huangfu D, Rudensky AY, Melnick AM, Leslie CS. Nature Communications 2024; PMID: 39487131
-
CRISPR screening uncovers a long-range enhancer for ONECUT1 in pancreatic differentiation and links a diabetes risk variant
-
3D Enhancer-promoter networks provide predictive features for gene expression and coregulation in early embryonic lineages
Murphy D, Salataj E, Di Giammartino DC, Rodriguez-Hernaez J, Kloetgen A, Garg V, Char E, Uyehara CM, Ee LS, Lee U, Stadtfeld M, Hadjantonakis AK, Tsirigos A, Polyzos A, Apostolou E. Nature Structural Molecular Biology. 2023 PMID: 38053013.
-
Dynamic network-guided CRISPRi screen identifies CTCF-loop-constrained nonlinear enhancer gene regulatory activity during cell state transitions
Luo R, Yan J, Oh JW, Xi W, Shigaki D, Wong W, Cho HS, Murphy D, Cutler R, Rosen BP, Pulecio J, Yang D, Glenn RA, Chen T, Li QV, Vierbuchen T, Sidoli S, Apostolou E, Huangfu D and Beer MA. Dynamic network-guided CRISPRi screen identifies CTCF-loop-constrained nonlinear enhancer gene regulatory activity during cell state transitions. Nature Genetics 2023; PMID: 37488417
-
3D enhancer-promoter interactions and multi-connected hubs: Organizational principles and functional roles
Chris M. Uyehara and Effie Apostolou, Cell Reports, 2023 Volume 42, Issue 4, 112068, ISSN 2211-1247, doi.org/10.1016/j.celrep.2023.112068.
-
A bipartite element with allele-specific functions safeguards DNA methylation imprints at the Dlk1-Dio3 locus
Aronson BE, Scourzic L, Shah V, Swanzey E, Kloetgen A, Polyzos A, Sinha A, Azziz A, Caspi I, Li J, Pelham-Webb B, Glenn RA, Vierbuchen T, Wichterle H, Tsirigos A, Dawlaty MM, Stadtfeld M* and Apostolou E*.
Developmental Cell 2021 Nov 22;56(22):3052-3065.e5. doi: 10.1016/j.devcel.2021.10.004.
-
OCT2 pre-positioning facilitates cell fate transition and chromatin architecture changes in humoral immunity
Doane AS, Chu CS, Di Giammartino DC, Rivas MA, Hellmuth JC, Jiang Y, Yusufova N, Alonso A, Roeder RG, Apostolou E, Melnick AM, Elemento O.
Nature Immunology 2021 Oct;22(10):1327-1340. doi: 10.1038/s41590-021-01025-w. PMID: 34556886
-
Deciphering the Complexity of 3D Chromatin Organization Driving Lymphopoiesis and Lymphoid Malignancies
Laurianne Scourzic, Eralda Salataj and Effie Apostolou
Frontiers in Immunology 2021 May 14;12:669881 PMID: 34054841
-
H3K27ac bookmarking promotes rapid post-mitotic activation of the pluripotent stem cell program without impacting 3D chromatin reorganization
Molecular Cell. 2021 Apr 15;81(8):1732-1748.e8. PMID: 33730542 -
Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture
Nevin Yusufova, Andreas Kloetgen, Matt Teater, Adewola Osunsade, Jeannie M Camarillo, Christopher R Chin, Ashley S Doane, Bryan J Venters, Stephanie Portillo-Ledesma, Joseph Conway, Jude M Phillip, Olivier Elemento, David W Scott, Wendy Béguelin, Jonathan D Licht, Neil L Kelleher, Louis M Staudt, Arthur I Skoultchi, Michael-Christopher Keogh, Effie Apostolou, Christopher E Mason, Marcin Imielinski, Tamar Schlick, Yael David, Aristotelis Tsirigos, C David Allis, Alexey A Soshnev, Ethel Cesarman, Ari M Melnick
Nature 2021 Jan;589(7841):299-305. PMID: 33299181
-
Identification of Cancer Drivers at CTCF Insulators in 1,962 Whole Genomes.
Cell Systems. 2019 May 22;8(5):446-455.e8. PMID: 31078526 -
Transcription factors: building hubs in the 3D space.
Cell Cycle. 2020 Oct;19(19):2395-2410. PMID: 32783593 -
KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks.
Nature Cell Biology. 2019 Oct;21(10):1179-1190. PMID: 31548608 -
A Susceptibility Locus on Chromosome 13 Profoundly Impacts the Stability of Genomic Imprinting in Mouse Pluripotent Stem Cells.
Cell Reports. 2020 Mar 17;30(11):3597-3604.e3. doi: 10.1016/j.celrep.2020.02.073.PMID: 32187532 -
Dynamic 3D Chromatin Reorganization during Establishment and Maintenance of Pluripotency.
Pelham-Webb B, Murphy D and Apostolou E.
Stem Cell Reports. 2020 Dec 8;15(6):1176-1195. PMID: 33242398
-
Context-Dependent Requirement of Euchromatic Histone Methyltransferase Activity during Reprogramming to Pluripotency.
Vidal SE, Polyzos A, Chatterjee K, Ee LS, Swanzey E, Morales-Valencia J, Wang H, Parikh CN, Amlani B, Tu S, Gong Y, Snetkova V, Skok JA, Tsirigos A, Kim, Apostolou E and Stadtfeld M.
Stem Cell Reports. 2020 Dec 8;15(6):1233-1245. PMID: 32976761
-
Wide-spread Mitotic Bookmarking by Histone Marks and Transcription Factors in Pluripotent Stem Cells.
Cell Reports. 2017 May 16;19(7):1283-1293. PMID: 28514649
-
Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming.
Cell Stem Cell. 2013 Jun 6;12(6):699-712. PMID: 23665121 -
Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells.
Nature. 2010 May 13;465(7295):175-81. PMID: 20418860 -
Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression.
Cell. 2008 Jul 11;134(1):85-96. PMID: 18614013
Press
-
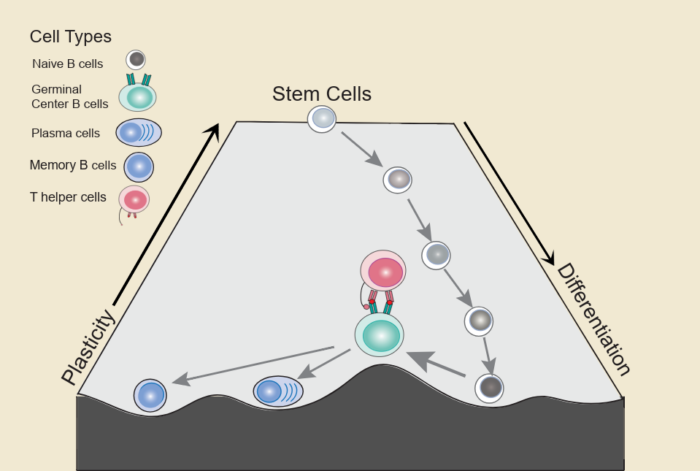
Article Featuring our work on the transient gain of plasticity during Germinal Center Reaction
This study, published Dec. 29 in Nature Cell Biology, reveals a paradox: as mature B cells get prepped to make antibodies, a highly specialized process, they temporarily gain plasticity, a feature normally reserved for unspecialized stem cells.
-
Weill Cornell Newsroom featuring our work on the role of 3D regulatory hubs on Glioblastoma
The findings, published April 3 in Molecular Cell, offer a new way to think about cancer beyond gene mutations, based on the way that genes are connected and regulated in three-dimensional space.
-
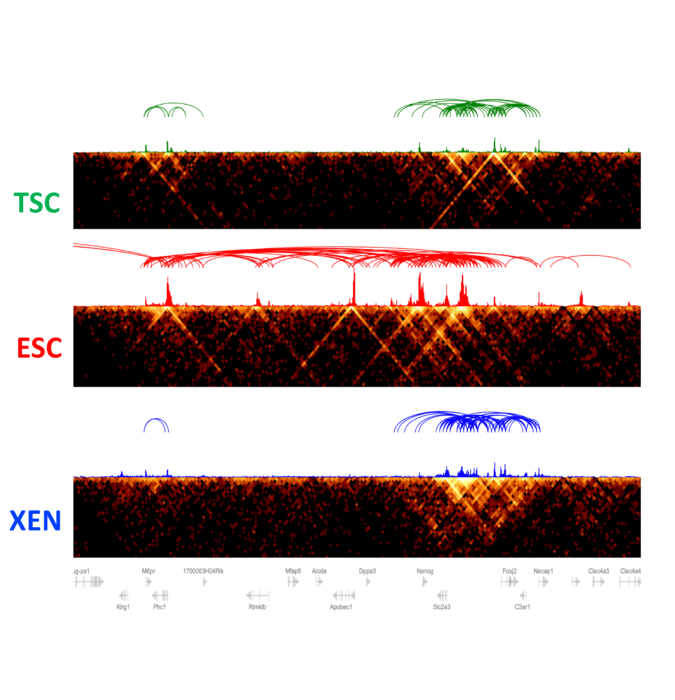
Article featuring our work on 3D regulatory networks in early embryonic decisions
n the nucleus of cells, long strands of DNA are tightly wrapped around a scaffolding of proteins in a complex called chromatin, like a rolled-up ball of yarn. A new study by Weill Cornell Medicine investigators reveals that beyond providing a convenient way to store DNA in a tight space, the 3-dimensional (3D) organization of noncoding gene regulators in chromatin contributes to the control of key cell identity programs in early embryonic development. The results have implications for understanding this critical period and how changes in the 3D chromatin architecture may contribute to developmental abnormalities or diseases like diabetes or cancer.
-
Article featuring our research on the molecular resetting of stem cell identity after cell division
Molecular "bookmarks," which allow cells to retain their characteristics during cell division, ensure fast reactivation of critical cell identity genes after cell division, according to investigators at Weill Cornell Medicine. The new work helps illuminate a process that has puzzled biologists for decades and suggests new strategies for modulating cell fate both for stem cell therapy and cancer treatment.
-
Article from Cornell Research featuring our recent work and publications.
Embryonic development starts from a single cell known as a zygote. Through multiple rounds of proliferation and differentiation, this initial cell generates the vast array of specialized cells that make up the body. But each time a cell proliferates and goes through mitosis—the process of cell division that results in two genetically identical daughter cells—it faces an identity crisis. It must decide whether to keep its original identity through self-renewal or give rise to a new one through differentiation.
-
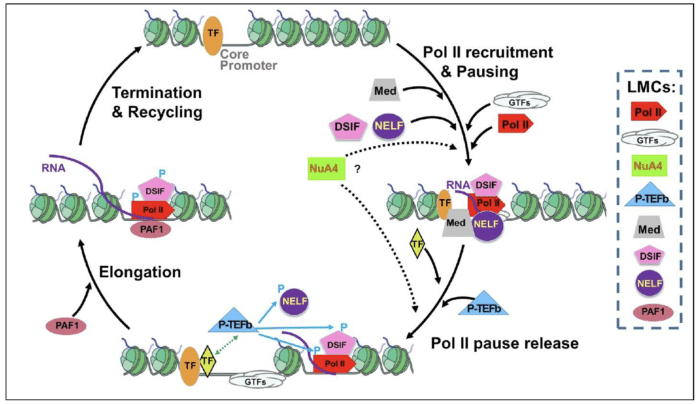
Multi-million NIH funding supports inter-campus collaboration between the Apostolou group and colleagues from Ithaca Cornell Campus
A Multi-PI team (Lis, Yu, Apostolou and Josefowicz) will use a bevy of state-of-the-art technologies to investigate large macromolecular complexes involved in transcription and its regulation. They developed an RNA aptamer – RNA selected to tightly bind to a green fluorescent protein – for tagging large complexes. The group will focus initially on RNA Polymerase II, the main machine that transcribes mRNA encoding genes.